|
Workunit: P000001 Title:modalign1  Go to: [Template Selection] [Alignment] [Modelling Log] [Evaluation] Print/Save this page as |
|||||||||||||||||||
Model Details:  Segment 1
Segment 1
Alignment  [top]
[top]
Anolea / Gromos / Verify3D  [top]
[top] 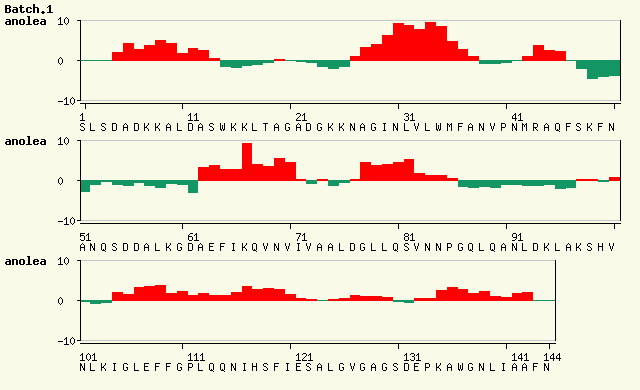 Modelling log  [top]
[top]3.70 (SP3) Loading Template: 1ux9A.pdb Loading Raw Sequence Loading Alignment: ./user.align.submit.fasta.FF Removing HET groups from template structure Refining Raw Sequence Alignment ProModII: doing simple assignment of backbone ProModII: adding blocking groups Adding Missing Sidechains AddPolar H BuildDeletetedLoopsModel Building CSP loop with anchor residues ASP 22 and ASN 26 Building CSP loop with anchor residues ALA 21 and ASN 26 Number of Ligations found: 80 ACCEPTING loop 62: clash= 0 FF= -13.4 PP= 2.00 Building CSP loop with anchor residues VAL 100 and LYS 103 Number of Ligations found: 5 ACCEPTING loop 4: clash= 0 FF= -69.0 PP= -2.00 Optimizing Sidechains Adding Hydrogens Optimizing loops and OXT (nb = 9) Final Total Energy: -5116.782 KJ/mol Dumping Sequence Alignment | |||||||||||||||||||
|
|
|||||||||||||||||||
If you publish results using SWISS-MODEL, please cite the following papers:
|
|||||||||||||||||||