Disease-causing mutations
It is useful to know where in the 3D structure of the protein the disease-causing mutations occur.
Viewing the structure
To visualize the 3D structure, click on the image in the 3D structures section.
You can now select one or more of the structures that you wish to view (superimposed on one another if you select more than one). We will only select one for this exercise. There are a number of viewing options, either using RasMol or JSMol which is simpler as it runs in your web browser.
- Select only the first structure which is a proper experimental
structure rather than one obtained from AlphaFold2 - its PDB
code is
7aak. - Select JSMol next to Display mode
- Next to Show: select all atoms
- click Display
A new tab will open showing the structure as a wireframe display with ligands shown as ball-and-stick.
We will now highlight some residues known to be mutated in the human protein. These are taken from the OMIM page under "ALLELIC VARIANTS" (they can also be found on the UniProtKB/Swiss-Prot page under "FT VARIANT").
Remember that the residue-numbers given in OMIM and UniProtKB/Swiss-Prot relate to the protein found in humans. If we were looking at a PDB file (or a model) from another species the residue numbering could well be different (owing to divergence of the two proteins from their common ancestor giving rise to insertions and deletions). Consequently, we would have to look at the alignment to work out the equivalent residue numbers in the PDB file that we are viewing.
Here we are looking at a structure of the human protein which has been numbered correctly despite the first few amino acids not being present in the PDB file
| Residue | Disease-causing mutation |
OMIM ref. no. |
|---|---|---|
| Arg26 | =>His | 0015 |
| Ala31 | =>Thr | 0016 |
| Gln34 | =>Lys | 0017 |
| Arg149 | =>Lys =>Gln |
- 0008 |
| Arg167 | =>Gln =>Trp =>Leu |
0005 0013 0014 |
We will now highlight these residues.
- Right-click on the JSMol image and select Console from the menu
- Enter the following commands into the console to highlight these residues and see where they occur in the 3D structure. (You can use your mouse to cut-and-paste them rather than typing them all in).
select *
colour cpk
select not protein and not water
spacefill
select Arg26
colour red
select Ala31
colour orange
select Gln34
colour green
select Arg149
colour purple
select Arg167
colour blue
define mres (Arg26 || Ala31 || Gln34 || Arg149 || Arg167)
select mres
wireframe 0.3
select mres & *.N
label " %n%r"
You should get something that looks like:
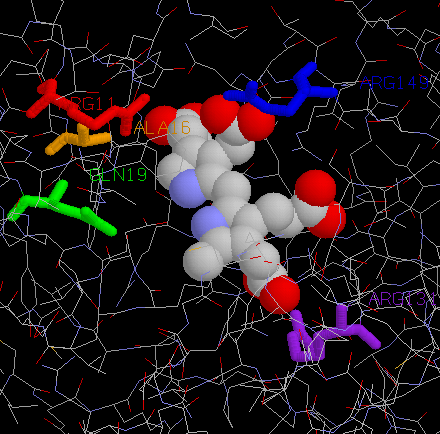
If you are not familiar with using JSMol, try moving the molecules around with the mouse (using a click-and-drag action within the JSMol window), as follows:
| Operation | Action |
|---|---|
| Left mouse button | Rotate about x-y |
| Ctrl-key + right mouse button | Translate x-y |
| Shift-key + left mouse button (move up and down) | Zoom in/out |
| Shift-key + left mouse button (move left and right) | Rotate about z |
The command language is almnost identical to that used in RasMol. For further information see the RasMol Introduction.
Consider the following:
From looking at the location of the above residues in the 3D structure, what can you say about their significance?