Submitting Your Own Alignment
In some cases, a 'better' template may be available which has been rejected by SwissModel. For example, a structure may be available which covers more of the target sequence, but has a lower overall sequence identity. In other cases, making changes to the sequence alignment may be important in order to improved the model.
Either way, we can use the 'Alignment Interface' mode of SwissModel to provide our own template(s) and alignment.
In the 'Alignment Interface' of the SwissModel page, you will see a text box into which you can paste an alignment. Here is hand-edited alignment with an alternative template structure, '1ux9A', shown in ClustalW format .
CLUSTAL W (1.83) multiple sequence alignment
TARGET SLSDADKKALDASWKKLTAGADGKKNAGINLVLWMFANVPNMRAQFSKFNANQSDDALKG
1ux9A ELSEAERKAVQAMWARLYANSE---DVGVAILVRFFVNFPSAKQYFSQFKHMEDPLEMER
.**:*::**::* * :* *.:: :.*: ::: :*.*.*. : **:*: :. ::
TARGET DAEFIKQVNVIVAALDGLLQSVNNPGQLQANLDKLAKSHVNLKIGLEFFGPLQQNIHSFI
1ux9A SPQLRKHASRVMGALNTVVENLHDPDKVSSVLALVGKAHA-LKHKVEPVYFKILSGVILE
..:: *:.. ::.**: :::.:::*.::.: * :.*:*. ** :* . .
TARGET ESALGVGAGSDEPKAWGNLIAAFN--------------
1ux9A VVAEEFASDFPPETQRAWAKLRGLIYSHVTAAYKEVGW
* .. . : :
This can simply be cut-and-paste into the server, but again we won't wait for the server, we will look at some pre-calculated results.
View the SwissModel output page for the second model.
Since we submitted our own alignment (and therefore implicitly specified which template structure should be used) the results page now only contains details of the modelling process itself. The various warnings may be useful if you want to look at the model in detail later. The most useful thing at the moment is the total energy score.
Record the reported energy for this new model.
Comparing the models
Based on the energies of the two models, think about which model is more likely to be correct, remembering that a lower energy (or more negative energy) indicates a more stable structure.
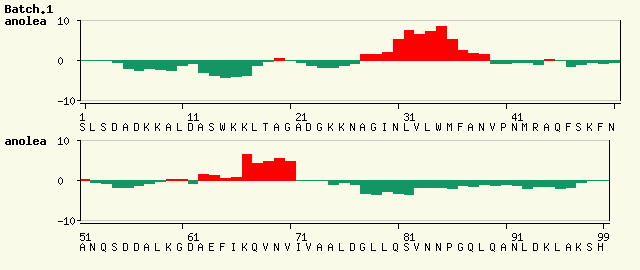
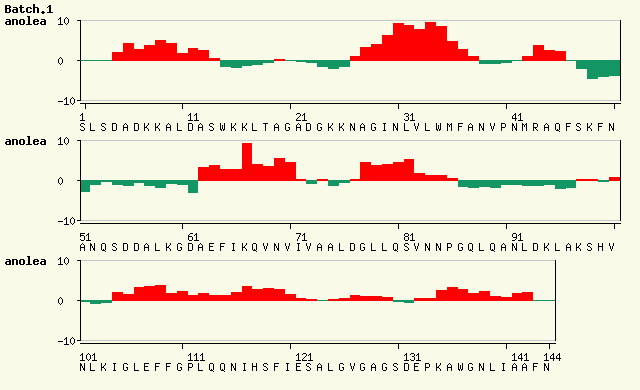
Now look at the ANOLEA results for the two models taken from the SwissModel results:
The ANOLEA results for the second model (Figure 2) are significantly worse than for the first model (Figure 1) - there are a lot more red regions in the graph. However, the template structure used for the first model (1it2B) has a resolution of 1.6Â while the template used for the second model (1ux9A) has a resolution of 2.4Â. Consequently the lower resolution of the template used for the second model may result in a poorer quality structure. We therefore need to look at the ANOLEA results for the template structures as well as for the resulting models to have a full picture of what is happening.