Least-squares fitting and RMSD
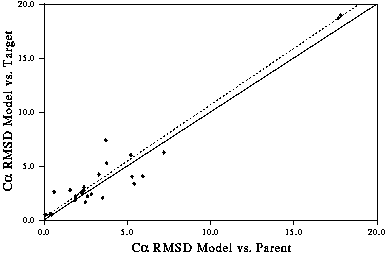
As shown in the graphs you looked at earlier, one indication of the quality of a model is how much you have had to change the template structure(s) to generate the model. i.e. The more changes made to the template(s), the more likely you are to make a mistake. This was shown in Figure 6 in the Introduction which is reproduced here.
Consequently, we will now compare the two models we have generated with their template structures using a program called ProFit. ProFit is a powerful and complex program for calculating the RMSD between protein structures. Here we will use a simple web interface which provides only the functionality we require.
Downloaded the two PDB models you generated by right-clicking on the links below and select Save Link As..., saving the files to your desktop or somewhere else memorable.
Now start the ProFit interface by clicking here.
Your first model was built using PDB Code 1it2, chain B
Your second model was built using PDB Code 1ux9, chain A
Record the Cα RMSD between the first model and its parent
Record the Cα RMSD between the second model and its parent